
Opinion: It’s time to stop product and science testing on animals
Mark Hawthorne
Science has given us many marvels. Electricity immediately springs to mind, as does the telephone. The remarkable ease with which we’re able to trace our ancestry is utterly astounding, and I have no doubt enjoyed more than my fair share of instant oatmeal. I am grateful to science for these and so many other life-changing wonders.
What I cannot abide, however, are scientific advances achieved through the pain and deaths of animals. '
Wednesday is World Day for Animals in Laboratories. On this day, animal advocates commemorate the suffering of millions of mice, rabbits, dogs, fishes, monkeys, pigs and other animals used in research, product testing and medical training. (The day is observed on April 24 to honor antivivisectionist Hugh Dowding, who was born in Scotland on this day in 1882.)
Humane Society International estimates that more than 115 million animals worldwide are used in laboratory experiments every year. These animals are beaten, blown up, burned and blinded. They are starved, suffocated, shaken and shot. They are nailed down, tied up and sliced open.
People are also reading…
They are forced to inhale tobacco smoke, drink alcohol and consume a variety of highly dangerous narcotics, including heroin. They are infected with diseases, sickened with toxic chemicals and injected with lethal pathogens. Their organs are pulverized, their limbs are severed, their bodies are irradiated and their spirits are broken. Nearly all these animals are killed after the experiment.
Animal testing is one of the most contentious practices in our society, pitting scientists and researchers against animal advocates and a growing population of compassionate consumers.
Since we share a large percentage of our genes with mice, chimpanzees, dogs and other vertebrates, scientists have commonly assumed that these animals are good models of human biology and that whatever harms them will also harm us.
The dangerous reality, however, is that there exist many disparities between the animal models and the human condition, and this translational inaccuracy can result in deadly failures. Four young boys perished after participating in a gene therapy trial in 2020, for instance, and three adults died in 2022 after taking an experimental drug for Alzheimer’s disease called lecanemab.
Indeed, according to the FDA, 92 percent of the drugs that prove safe and therapeutically effective in animals fail in clinical trials using humans. One study published by the National Institutes of Health (NIH), the world’s largest funder of biomedical research, found that of 93 dangerous drug side effects, only 19 percent could have been predicted by studies using animals.
I can already hear the “Yes, but …” defenses: Animal experiments save so many human lives; we can’t find cures without animal testing; animal models give us medical advances and promising treatments; et cetera.
It is true that animal testing has led to benefits for humans; it would be disingenuous to suggest otherwise. But there are humane alternatives to animal testing that do not require the exploitation of animals.
In February, Monica M. Bertagnolli, director of the NIH, announced that they plan to prioritize “novel alternative methods” of research that do not rely on animal testing. “These complementary, non-animal-based approaches hold tremendous promise,” she wrote on X.
Such alternatives include in vitro testing, which involves conducting experiments in controlled laboratory environments using human cells or tissues or sophisticated cell culture systems; organoids, which are three‐dimensional miniaturized versions of organs or tissues that are derived from cells; organ-on-a-chip technology, an in-vitro micro-scale biomimetic platform that helps in reproducing physiological environment of human organs; and artificial intelligence, which can analyze vast data from computational models and predict drug interactions.
And let’s not overlook the role of human volunteers in testing the safety and efficacy of new treatments or products. Micro-dosing, for instance, involves administering small drug doses to volunteers, affecting cells without causing major physiological effects and helping to identify nonviable drugs.
We have a moral obligation to animals. The NIH’s announcement — coupled with the passage in 2022 of the FDA Modernization Act 2.0 allowing drug developers to use non-animal methods to test for safety — gives me hope that humanity is edging ever closer to a day when we are no longer subjecting these sentient beings to pain, emotional suffering and death in laboratories.
Please contact your members of Congress and urge them to support the Humane Cosmetics Act, H.R. 5399, legislation that would prohibit animal testing for cosmetics manufactured or sold in the U.S. Click here for more information.
Hawthorne serves on the board of Save the Buns, a nonprofit that provides sanctuary and finds loving homes for rabbits who have been rescued from product-testing and research labs, and is the author of "Bleating Hearts: The Hidden World of Animal Suffering."
Get opinion pieces, letters and editorials sent directly to your inbox weekly!
Source: Opinion: It’s time to stop product and science testing on animals

Retractions are part of science, but misconduct isn’t — lessons from a superconductivity lab
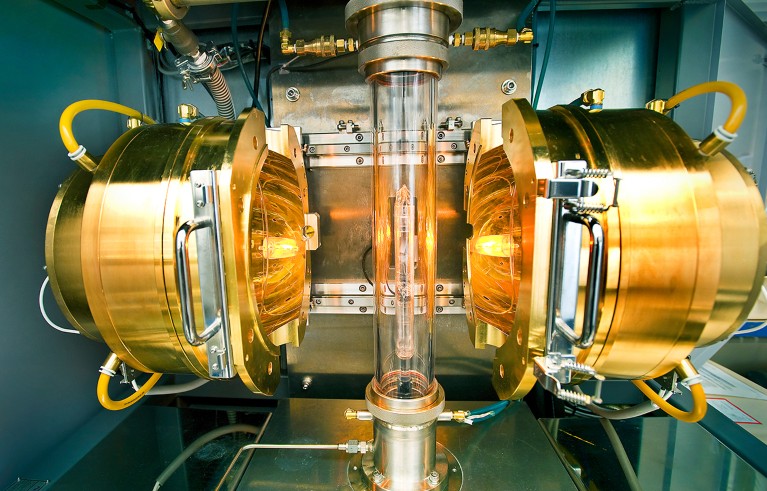
Superconductivity has been demonstrated at extremely low temperatures, but it remains elusive at room temperatures.Credit: Brookhaven National Laboratory/SPL
Research misconduct is hugely detrimental to science and to society. Defined as “fabrication, falsification, or plagiarism in proposing, performing, or reviewing research, or in reporting research results” by the US Office of Research Integrity, it violates trust in science and can do great harm to the wider public, scientific institutions and especially co-authors and students who had no part in the wrongdoing. In cases involving public funds, it squanders resources that could have been allocated to other research and it can erode lawmakers’ support for science.
Does the scientific community, as a whole, have appropriate processes for reporting, investigating and communicating about instances of potential misconduct? This question is not new. At Nature, we’re asking it again, after two separate studies that we published were subsequently retracted.
Exclusive: official investigation reveals how superconductivity physicist faked blockbuster results
The studies1,2 were originally published in October 2020 and March 2023. The first was retracted in September 2022 and the second in November 2023. The corresponding author on both papers was Ranga Dias, a physicist studying superconductivity at the University of Rochester in New York, and a recipient of grants from the US National Science Foundation (NSF).
The papers by Dias and his co-authors claimed to report room-temperature superconductivity under extremely high pressures, each in different materials. Room-temperature superconducting materials are highly sought after. They could, for example, transform the efficiency of electricity transmission, from the smallest to the largest application. But high-pressure experiments are difficult and replicating them is complex.
Nature initiated an investigative process that resulted in the 2020 paper being retracted after members of the community told the journal they were troubled by aspects of the data being reported. Nature also initiated an investigation into the 2023 paper. However, this article was retracted at the request of most of Dias’s co-authors while the investigation was still ongoing.
Many details about this case came to light thanks to continued questions from the research community, including during post-publication peer review. Much credit must also go to the persistence of science journalists, including members of Nature’s news team (which is editorially independent of Nature’s journal team) and those from other publications.
What can journal editors, funding organizations and institutions that employ researchers learn from such cases? We have the same goal: producing and reporting rigorous research of the highest possible standard. And we need to learn some collective lessons — including on the exchange of information.
The University of Rochester conducted three inquiries, which are a preliminary step to making a decision about whether to perform a formal investigation into scientific misconduct. The inquiries were completed between January and October 2022. Each concluded that such an investigation was not warranted.

Superconductivity scandal: the inside story of deception in a rising star’s physics lab
Earlier this month, Nature’s news team uncovered a 124-page report on a subsequent confidential investigation, performed at the NSF’s request. In it, a team of reviewers concluded after a ten-month assessment of evidence that it was more likely than not that Dias had committed data fabrication, falsification and plagiarism. The report is dated 8 February 2024, and the determination is regarding the two Nature papers, a 2021 study3 published in Physical Review Letters and a 2022 study4 in Chemical Communications — both of which were also retracted. However, the investigation has not yet officially been made public.
Some researchers have asked why Nature published Dias’s second paper in March 2023, when questions were being asked about the first one. Others have asked why the retraction notices didn’t spell out that there has been misconduct.
It’s important to emphasize that it’s Nature’s editorial policy to consider each submission in its own right. Second, peer review is not designed to identify potential misconduct. The role of a journal in such situations is to correct the scientific literature; it is for the institutions involved to determine whether there has been misconduct, and to do so only after the completion of due process, which involves a systematic evaluation of primary evidence, such as unmodified experimental data.
Access to raw data is fundamental to resolving cases of potential misconduct. It is also something we constantly think about in relation to publishing. Indeed, for certain kinds of data, Nature requires authors to deposit them in external databases before publication. But there must be more the research community — including funders and institutions — can all do to incentivize data sharing.
Another question is whether the matter could have been dealt with more quickly. Nature’s editors have been asking the same question: specifically, could there have been more, or better, communication between journals and institutions once evidence of potential misconduct came to light?

Publish, and be damned...
Last month, the Committee on Publication Ethics (COPE), a non-profit organization that represents editors, publishers and research institutions, updated its guidelines on how publishers and universities could communicate better. The guidelines are full of important advice, including that institutions, not publishers, should perform integrity or misconduct investigations. Investigators require access to primary evidence. As employers and grant-givers, institutions are the appropriate bodies to mandate access to unmodified experimental data, correspondence, notebooks and computers and to interview relevant staff members — all essential parts of an investigation.
But often, journals need to start a process that could lead to retracting a study in the absence of an institutional investigation — or while an investigation, or inquiry, is ongoing5. Are cases such as this an opportunity for journals and institutions to discuss establishing channels through which to exchange information, in the interest of expedited outcomes — as part of due process? Nature’s editors would be willing to play a part in such discussions.
Retractions are part of publishing research, and all journals must be committed to retracting papers after due process is completed. Although a paper can be retracted for many reasons, when the cause is potential misconduct, institutions must conduct thorough investigations.
This case is not yet closed. Both the university and the funder need to formally announce the investigation’s results, and what action they intend to take. They should not delay any more than is necessary. When there is credible evidence of potential scientific misconduct, investigations should not be postponed. There is strength in collaborating to solve a problem, and nothing to be ashamed of in preserving the integrity of the scientific record.
Source: Retractions are part of science, but misconduct isn’t — lessons from a superconductivity lab

Vast DNA tree of life for plants revealed by global science team using 1.8 billion letters of genetic code
A new paper published today (April 24) in the journal Nature by an international team of 279 scientists led by the Royal Botanic Gardens, Kew presents the most up-to-date understanding of the flowering plant tree of life.
Using 1.8 billion letters of genetic code from more than 9,500 species covering almost 8,000 known flowering plant genera (ca. 60%), this incredible achievement sheds new light on the evolutionary history of flowering plants and their rise to ecological dominance on Earth.
The study's authors believe the data will aid future attempts to identify new species, refine plant classification, uncover new medicinal compounds, and conserve plants in the face of climate change and biodiversity loss.
The major milestone for plant science, led by Kew and involving 138 organizations internationally, was built on 15 times more data than any comparable studies of the flowering plant tree of life. Among the species sequenced for this study, more than 800 have never had their DNA sequenced before.
The sheer amount of data unlocked by this research, which would take a single computer 18 years to process, is a huge stride towards building a tree of life for all 330,000 known species of flowering plants—a massive undertaking by Kew's Tree of Life Initiative.
Dr. Alexandre Zuntini, Research Fellow at RBG Kew, says, "Analyzing this unprecedented amount of data to decode the information hidden in millions of DNA sequences was a huge challenge. But it also offered the unique opportunity to reevaluate and extend our knowledge of the plant tree of life, opening a new window to explore the complexity of plant evolution."
Unlocking historic herbarium specimens for cutting-edge research
The flowering plant tree of life, much like our own family tree, enables us to understand how different species are related to each other. The tree of life is uncovered by comparing DNA sequences between different species to identify changes (mutations) that accumulate over time like a molecular fossil record.
Our understanding of the tree of life is improving rapidly in tandem with advances in DNA sequencing technology. For this study, new genomic techniques were developed to magnetically capture hundreds of genes and hundreds of thousands of letters of genetic code from every sample, orders of magnitude more than earlier methods.
A key advantage of the team's approach is that it enables a wide diversity of plant material, old and new, to be sequenced, even when the DNA is badly damaged. The vast treasure troves of dried plant material in the world's herbarium collections, which comprise nearly 400 million scientific specimens of plants, can now be studied genetically.
Using such specimens, the team successfully sequenced a sandwort specimen (Arenaria globiflora) collected nearly 200 years ago in Nepal and, despite the poor quality of its DNA, were able to place it in the tree of life.
The team even analyzed extinct plants, such has the Guadalupe Island olive (Hesperelaea palmeri), which has not been seen alive since 1875. In fact, 511 of the species sequenced are already at risk of extinction, according to the IUCN Red List, including three more like Hesperelaea that are already extinct.
Professor William Baker, Senior Research Leader–Tree of Life, says, "In many ways this novel approach has allowed us to collaborate with the botanists of the past by tapping into the wealth of data locked up in historic herbarium specimens, some of which were collected as far back as the early 19th century.
"Our illustrious predecessors such as Charles Darwin or Joseph Hooker could not have anticipated how important these specimens would be in genomic research today. DNA was not even discovered in their lifetimes!
"Our work shows just how important these incredible botanical museums are to ground-breaking studies of life on Earth. Who knows what other undiscovered science opportunities lie within them?"
Across all 9,506 species sequenced, more than 3,400 came from material sourced from 163 herbaria in 48 countries. Additional material from plant collections around the world (e.g., DNA banks, seeds, living collections) have been vital for filling key knowledge gaps to shed new light on the history of flowering plant evolution. The team also benefited from publicly available data for more than 1,900 species, highlighting value of the open science approach to future genomic research.
Flowering plants alone account for about 90% of all known plant life on land and are found virtually everywhere on the planet—from the steamiest tropics to the rocky outcrops of the Antarctic Peninsula. And yet, our understanding of how these plants came to dominate the scene soon after their origin has baffled scientists for generations, including Charles Darwin.
Flowering plants originated more than 140 million years ago after which they rapidly overtook other vascular plants including their closest living relatives—the gymnosperms (non-flowering plants that have naked seeds, such as cycads, conifers, and ginkgo).
Darwin was mystified by the seemingly sudden appearance of such diversity in the fossil record. In an 1879 letter to Joseph Dalton Hooker, his close confidant and Director of RBG Kew, he wrote, "The rapid development as far as we can judge of all the higher plants within recent geological times is an abominable mystery."
Utilizing 200 fossils, the authors scaled their tree of life to time, revealing how flowering plants evolved across geological time. They found that early flowering plants did indeed explode in diversity, giving rise to more than 80% of the major lineages that exist today shortly after their origin.
However, this trend then declined to a steadier rate for the next 100 million years until another surge in diversification about 40 million years ago, coinciding with a global decline in temperatures. These new insights would have fascinated Darwin and will surely help today's scientists grappling with the challenges of understanding how and why species diversify.
A truly global collaboration
Assembling a tree of life this extensive would have been impossible without Kew's scientists collaborating with many partners across the globe. In total, 279 authors were involved in the research, representing many different nationalities from 138 organizations in 27 countries. They include the Genomics for Australian Plants (GAP) consortium who were early adopters of the team's techniques and who worked in close collaboration with Kew to maximize the number of Australian plant species in the tree.
International collaborators also shared their unique botanical expertise, as well as many precious plant samples from around the world that could not be obtained without their help. The comprehensive nature of the tree is in no small part a result of this wonderful partnership.
Dr. Mabel Lum, Program Manager at Bioplatforms Australia and from the GAP consortium, says, "We are proud to be a major partner and collaborator in RBG Kew's effort to build global research infrastructure to advance our understanding of flowering plant tree of life. This fruitful collaboration underscored our commitment to fostering innovation and collaboration in scientific research, providing a springboard for future discoveries that will help shape our understanding of the natural world for generations to come."
Putting the plant tree of life to good use
The flowering plant tree of life has enormous potential in biodiversity research. This is because, just as one can predict the properties of an element based on its position in the periodic table, the location of a species in the tree of life allows us to predict its properties. The new data will thus be invaluable for enhancing many areas of science and beyond.
To enable this, the tree and all of the data that underpin it have been made openly and freely accessible to both the public and scientific community, including through the Kew Tree of Life Explorer. The study's authors believe such open access is key to democratizing access to scientific data across the globe.
Open access will also help scientists to make the best use of the data, such as combining it with artificial intelligence to predict which plant species may include molecules with medicinal potential. Similarly, the tree of life can be used to better understand and predict how pests and diseases are going to affect the plants of the U.K. in the future. Ultimately, the authors note, the applications of this data will be driven by the ingenuity of the scientists accessing it.
Dr. Melanie-Jayne Howes, Senior Research Leader at RBG Kew who was not an author on the study but will make use of the data in her research, says, "Plant chemicals have inspired many pharmaceutical drugs, but still have great untapped potential to aid future drug discovery. The challenge is knowing which to investigate scientifically in the search for new medicines out of the ca. 330,000 flowering plant species.
"At Kew we are applying AI to predict which plant species contain chemicals with pharmaceutical potential for malaria. The availability of this vast new dataset offers exciting opportunities to enhance these predictions and hence accelerate drug discovery from plants for malaria and other diseases too."
Remarkable species in the flowering plant tree of life
More information: Zuntini, A. R., Carruthers, T. et al, Phylogenomics and the rise of the angiosperms, Nature (2024). www.nature.com/articles/s41586-024-07324-0
Citation: Vast DNA tree of life for plants revealed by global science team using 1.8 billion letters of genetic code (2024, April 24) retrieved 24 April 2024 from https://phys.org/news/2024-04-vast-dna-tree-life-revealed.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.
Source: Vast DNA tree of life for plants revealed by global science team using 1.8 billion letters of genetic code
